|
Witcher Lab members (left – right): Su Jie (Research Assistant), Dr. Sofiane Berrazouane (postdoctoral fellow), Meredyth Elisseou (graduate student), Mariam Anwar (graduate student), Eduardo Cepeda Canedo (graduate student), Emmanuel Asante (graduate student), Marios Langke (graduate student), Dr. Antoine Meant (postdoctoral fellow), Audrey Houle (graduate student), Dr. Michael Witcher (PI), Dr. Tiejun Zhao (Senior Research Assistant).
General Overview of Our Research:
Epigenetics is defined as molecular factors that regulate regulate genome activity, independently of DNA sequence. This includes histone modification, DNA methylation and non-coding RNAs. In our lab we study epigenetic defects in Breast and Ovarian Cancers. Through understanding how such aberrations give rise to cancer, we can gain critical insights allowing us to develop new therapeutic approaches to treat these diseases.
We incorporate both classical and cutting edge technologies to examine genome wide epigenetic reprogramming in cancer and develop new small molecule inhibitors targeting epigenetic processes. These techniques include, Chromatin-IP, ChIP-seq, RNA-seq, laser micro-irradiation, Mass Spectrometry, CRISPR-Cas9, and animal models of cancer.
 |
We think that cancer cells develop a signaling network connecting the micro-environment to chromatin regulators, resulting in epigenetic reprogramming. |
Specific Projects in the lab
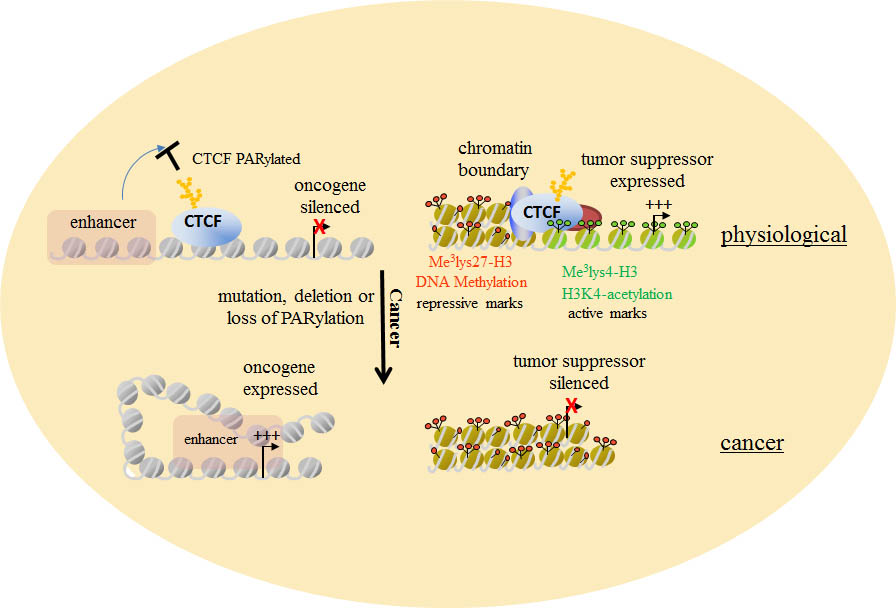
1. Understanding the consequences of CTCF dysfunction in cancer
 CTCF is a multi-functional, master- regulator of the human genome. It may be functionally inactivated in cancer via several mechanisms including; 1) Deletion 2) Mutation
3) loss of the post-translational modification poly (ADP-ribosylation) 4) Mutation of its DNA binding sites. CTCF is a multi-functional, master- regulator of the human genome. It may be functionally inactivated in cancer via several mechanisms including; 1) Deletion 2) Mutation
3) loss of the post-translational modification poly (ADP-ribosylation) 4) Mutation of its DNA binding sites.
We aim to explore the epigenetic and transcriptional consequences of these defects. Unexpectedly, we have established a new role for CTCF in the repair of double strand breaks. We are now exploring whether this novel function of CTCF is disrupted in cancer, and promotes oncogenic progression.
The transcription factor TFII-I targets CTCF to metabolism-related genes.
Results of ChIP-seq experiments.
2. Drugging the epigenome as an approach to fight ovarian cancer.
Using RNA-seq and Chromatin-IP, we have uncovered epigenetic defects which are enabling us to find new therapeutic avenues to treat untreatable cancers.
 |
RNA-seq data from Ovarian cancer cell lines treated with drugs targeting the epigenome.
|
3. Targeting poly (ADP-ribosylation)
Poly (ADP-ribosylation) is a protein modification placed by PARP-1. We have evidence that this pathway is abnormal in a spectrum of cancers. By targeting this pathway through unique means developed by my lab, we are able to either promote, or effectively slow, the growth of primary and metastatic breast cancers.
 |
Ablation of lung metastases by manipulating the poly(ADP-ribose) pathway.
|
 |
Triple-negative Breast Cancer cells reprogrammed to acquire more physiological, epithelial-like characteristics after modulation of the poly (ADP-ribose) pathway. |
Witcher Lab publications:
(* denotes corresponding authorship)
1) Lebeau B, Jangal M, Zhao T, Wong CK, Wong N, Cañedo EC, Hébert S, Aguilar-Mahecha A, Chabot C, Buchanan M, Catterall R, McCaffrey L, Deblois G, Kleinman C, Park M, Basik M, Witcher M*. 3D chromatin remodeling potentiates transcriptional programs driving cell invasion. Proc Natl Acad Sci., Sept 6, 2022 (IF 12.7)
2) Lebeau B, Zhao K, Jangal M, Zhao T, Guerra M, Greenwood CMT, Witcher M*. Single base-pair resolution analysis of DNA binding motif with MoMotif reveals an oncogenic function of CTCF zinc-finger 1 mutation. Nucleic Acids Res, Aug 6, 2022 (IF 19.1)
3) Heath J, Simo Cheyou E, Findlay S, Luo VM, Pinedo Carpio E, Lee J, Djerir B, Chen X, Morin T, Lebeau B, Karam M, Bagci H, Grapton D, Ursini-Siegel J, Côté JF, Witcher M, Richard S, Maréchal A, and Orthwein A. POGZ modulates the DNA damage response in a HP1-dependent manner. EMBO Rep., Nov 10 2021, e51041 (IF 8.8)
4) Totten SP, Im YK, Cepeda Cañedo E, Najyb O, Nguyen A, Hébert S, Ahn R, Lewis K, Lebeau B, La Selva R, Sabourin V, Martínez C, Savage P, Kuasne H, Avizonis D, Santos Martínez N, Chabot C, Aguilar-Mahecha A, Goulet ML, Dankner M, Witcher M, Petrecca K, Basik M, Pollak M, Topisirovic I, Lin R, Siegel PM, Kleinman CL, Park M, St-Pierre J, Ursini-Siegel J. STAT1 potentiates oxidative stress revealing a targetable vulnerability that increases phenformin efficacy in breast cancer. Nat Commun. 2021 Jun 3;12(1):3299. (IF 17.6)
5) Cañedo EC, Totten S, Ahn R, Savage P, MacNeil D, Hudson J, Autexier C, Deblois G, Park M, Witcher M*, Ursini-Siegel J (Co-corresponding). p66ShcA potentiates the cytotoxic response of triple-negative breast cancers to PARP inhibitors. JCI Insight. 2021 Feb 22;6(4):e138382 (IF 9.4)
6) Shorstova T, Foulkes WD, Witcher M Achieving clinical success with BET inhibitors as anti-cancer agents. Br J Cancer. 2021 Mar 15. doi: 10.1038/s41416-021-01321-0 (IF 7.6)
7) Tawil N, Bassawon R, Meehan B, Nehme A, Montermini L, Gayden T, De Jay N, Spinelli C, Chennakrishnaiah S, Choi D, Adnani L, Zeinieh M, Jabado N, Kleinman CL, Witcher M, Riazalhosseini Y, Key NS, Schiff D, Grover SP, Mackman N, Couturier CP, Petrecca K, Suvà ML, Patel A, Tirosh I, Najafabadi H, Rak J Glioblastoma cell populations with distinct oncogenic programs release podoplanin as procoagulant extracellular vesicles. Blood Adv. 2021 Mar 23;5(6):1682-1694. (IF 7.6)
8) Shorstova T, Su J, Zhao T, Dahabieh M, Leibovitch M, De Sa Tavares Russo M, Avizonis A, Rajkumar S, Watson I, del Rincon SV, Miller WH JR., Foulkes WD Witcher M*. Reprogramming of nucleotide metabolism mediates synergy between epigenetic therapy and MAP Kinase inhibition. Molecular Cancer Therapeutics, 2021, Jan;20(1):64-75. (IF 6.2)
9) Spriano F, Gaudio E, Cascione L, Tarantelli C, … Witcher M, Brown B, Wahlestedt C, Giles F, Stathis A and Bertoni F. Anti-tumor activity of the dual BET and CREBBP/EP300 inhibitor NEO2734. Blood Advances, 2020, Sep 8;4(17):4124-4135 (IF 7.6)
10) Tischkowitz M, Huang S, Banerjee S, Hague J, Hendricks WPD, Huntsman DG, Lang JD, Orlando KA, Oza AM, Pautier P, Ray-Coquard I, Trent JM, Witcher M, Witkowski L, McCluggage WG, Levine DA, Foulkes WD, Weissman BE. Clin Cancer Res. 2020 Aug 1;26(15):3908-3917 (IF 13.8)
11) Roy DG, Chen J, ... Witcher M, Krawczyk CM, Larochelle C, Jones RG. Methionine Metabolism Shapes T Helper Cell Responses through Regulation of Epigenetic Reprogramming. Cell Metabolism, 2020, Feb 4;31(2):250-266. (IF 27.2)
12) Jangal M, Lebeau B, Witcher M*. Beyond EZH2: is the polycomb protein CBX2 an emerging target for anti-cancer therapy? Expert Opin Ther Targets. 2019 Jul;23(7):565-578. (IF 5.2)
13) Shorstova T, Maud Marques M, Su J, Johnston J, Kleinman CL, Hamel N, Huang S, Alaoui-Jamali MA, Foulkes WD and Witcher M*. “Susceptibility of SMARCA4 deficient cancers to bromodomain inhibitors”. Cancer Research, 2019 May 15;79(10):2761-2774 (IF 12.7)
14) Guo Q, Li VZ, ...Amant F, Witcher M, Behbod F, McCaffrey L, Alaoui-Jamali MA, Giannakopoulos NV, Brackstone M, Postovit LM, Del Rincón SV, Miller WH. MNK1/NODAL signaling promotes invasive progression of breast ductal carcinoma in situ. Cancer Research 2019, Apr 1;79(7):1646-1657 (IF 12.7)
15) Xue Y, Meehan B, Macdonald E, Venneti S, Wang XQ, Witkowski L, Kong T, Martinez D, Morin G, Abedini A, Johnson RM, Cencic R, Chen H, Jelinic P, Papadakis A, Auguste A, de Rink I, Kerkhoven RM, Bertos N, Gotlieb WH, Clarke BA, Leary A, Witcher M, Guiot MC, Pelletier J, Dostie J, Park M, Levine DA, Judkins AR, Hass R, Rak J, Vanderhyden B, Foulkes WD and Huang S. “CDK4/6 inhibitors target SMARCA4-determined cyclin D1 deficiency in small cell carcinoma of the ovary, hypercalcemic type”. Nature Communications, Feb 4;10(1):558. (IF 14.9)
16) “Oncogenic activity of Poly (ADP-ribose) glycohydrolase”. Marques M, Jangal M, Wang LC, Kazanets A, da Silva SD, Zhao T, Lovato A, Yu H, Jie S, Del Rincon S, Mackey J, Damaraju S, Alaoui-Jamali M and Witcher M*. Oncogene, Mar;38(12):2177-2191. (IF 8.7)
17) Wang Z, Wang W, Cui YC, Pan Q, Zhu W, Gendron P, Guo F, Cen S, Witcher M, Liang C. “HIV-1 employs multiple mechanisms to resist Cas9/sgRNA targeting the viral primer binding site.” J Virol. 2018 Aug 1. pii: JVI.01135-18 (IF 4.5)
18) Dahabieh MS, Di Pietro E, Jangal M, Goncalves C, Witcher M, Braverman NE, Del Rincón SV. “Peroxisomes and Cancer: The Role of a Metabolic Specialist in a Disease of Aberrant Metabolism.” BBA reviews Cancer. 2018 Jul 13. pii: S0304-419X(18)30053-2. (IF 10.6)
19) Tang L, Morris J, Wan J, Moore C, Fujita Y, Gillaspie S, Aube E, Nanda J, Marques M, Jangal M, Anderson A, Cox C, Hiraishi H, Dong L, Saito H, Singh CR, Witcher M, Topisirovic I, Qian SB, Asano K. “Competition between translation initiation factor eIF5 and its mimic protein 5MP determines non-AUG initiation rate genome-wide.” Nucleic Acids Res. 2017 Sep 18. doi: 10.1093/nar/gkx808 (IF 16.4)
20) Hilmi K, Jangal M, Marques M, Zhao T, Saad S, Zhang C, Luo VM, Syme A, Rejon C, Yu Z, Krum A, Fabian MR, Richard S, Alaoui-Jamali M, Orthwein A, McCaffrey L and Witcher M*. “CTCF facilitates DNA double-strand break repair by enhancing homologous recombination repair.” Science Advances (AAAS, Science). 2017 May 24;3(5):e1601898. (IF 13.1)
|